Multi-Omics-supported Personalized Treatment Recommendation for Patients
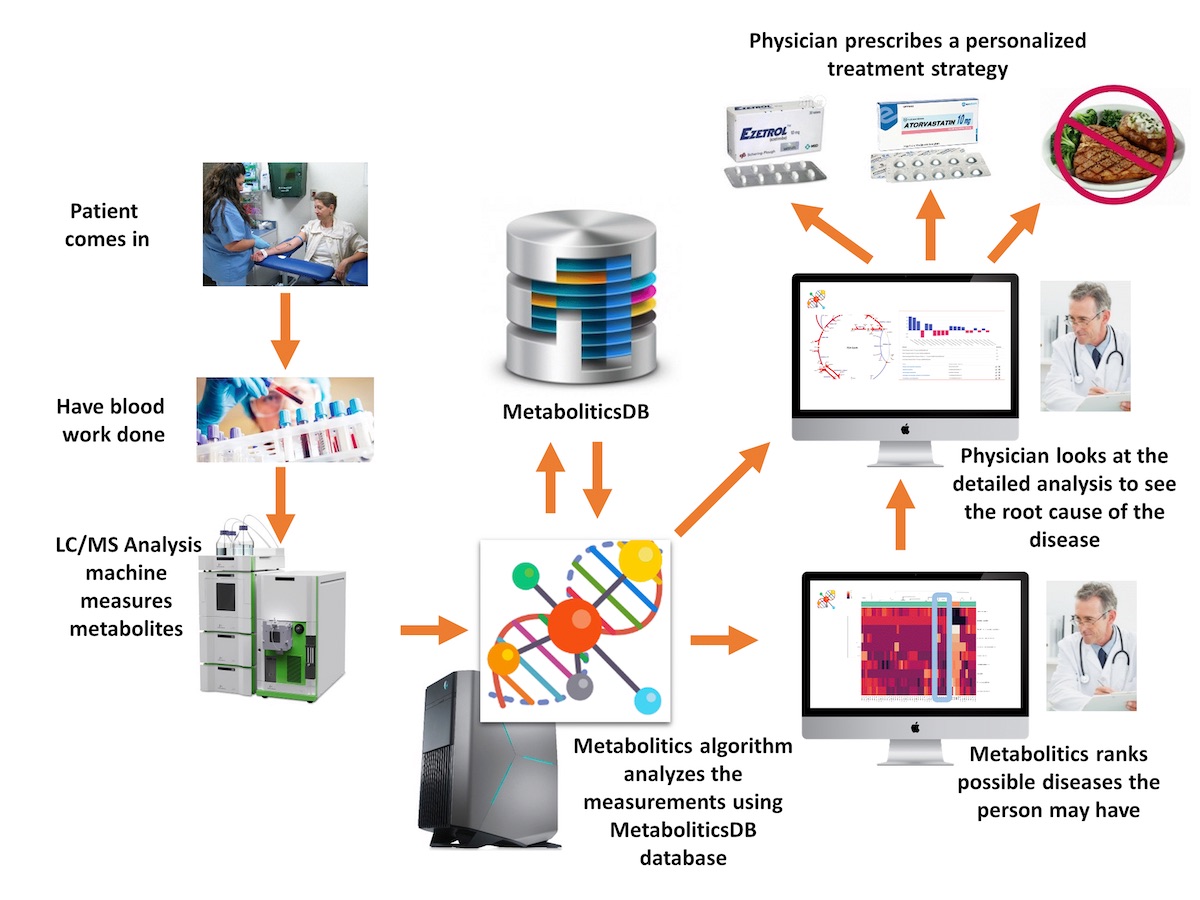
Increasing availability of high throughput omics data at decreasing costs has led to rapid progress in understanding the mechanisms of diseases. Such developments offer many opportunities for personalized and precision medicine practices. This project focuses on the development of tools and methods for the exploitation of the computed metabolism changes for personalized treatment selection (e.g., individualized drug recommendations for a patient). To this end, we will intensively use the machine learning models that will be built on the metabolic analysis results. In more detail, this research involves the following parts:
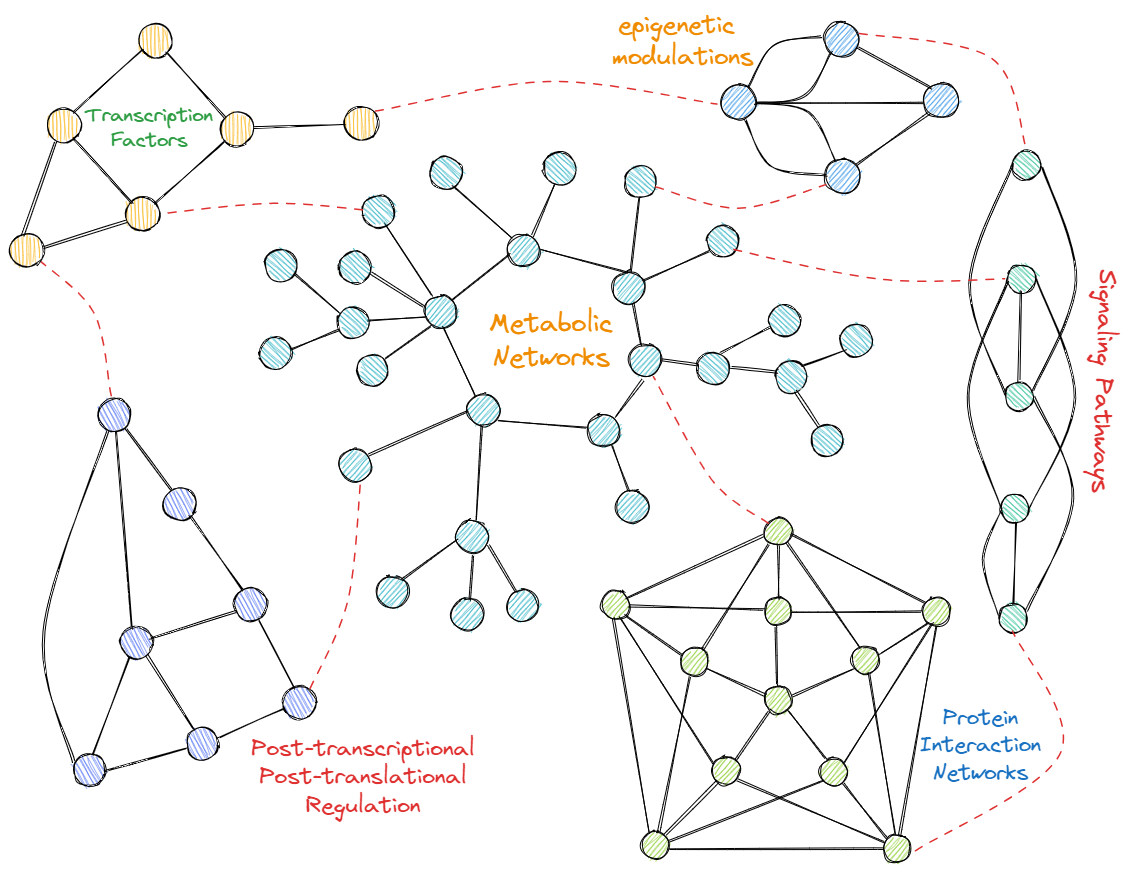
- Integrating transcriptome, proteome, and metabolome for more informed metabolic profile prediction: Metabolic networks are controlled by enzymes of the underlying processes and the regulators (i.e., activator, inhibitors, etc.) of those enzymes. Enzymes being mostly proteins (and, rarely, rRNAs), transcriptome and proteome of an organism are indirectly related to the metabolome of an organism. Hence, investigating effective integration strategies among different “ome”s (i.e., proteome, metabolome, and transcriptome) is vital to deriving more accurate metabolic profile predictions. We will extend our own Metabolitics algorithm to utilize multiple types of omics datasets to compute the metabolic profiles of patients.
-
Developing machine learning-based classification models for diseases: Through the metabolic changes calculated based on the metabolic analysis of omics data, machine learning models will be built to classify individuals as healthy or having a disease of interests. As datasets, databases from public omic data sources such as Metabolomics Workbench, TCGA, GEO, ProteomeXchange may be used.
-
Designing and creating a classification model database: Once the machine learning models are created, they will be used repeatedly at many points. Since the training and optimization of these models require time and resources, the reuse of the created models is important for the efficient use of resources. In this context, a classification model database may be designed and created to enable the reuse of the models as well as the efficient management and storage of the classification models created for diseases.
-
Developing an individualized drug recommendation system: In this step, a clinical decision support system will be developed to compute the most suitable treatment for a patient by blocking the target of each drug in the metabolic network of the patient and re-computing the metabolic profile of the patient under this constraint. The resulting profile will be fed into the classification model once again to check if the predicted class of the patient turns into “healthy”.