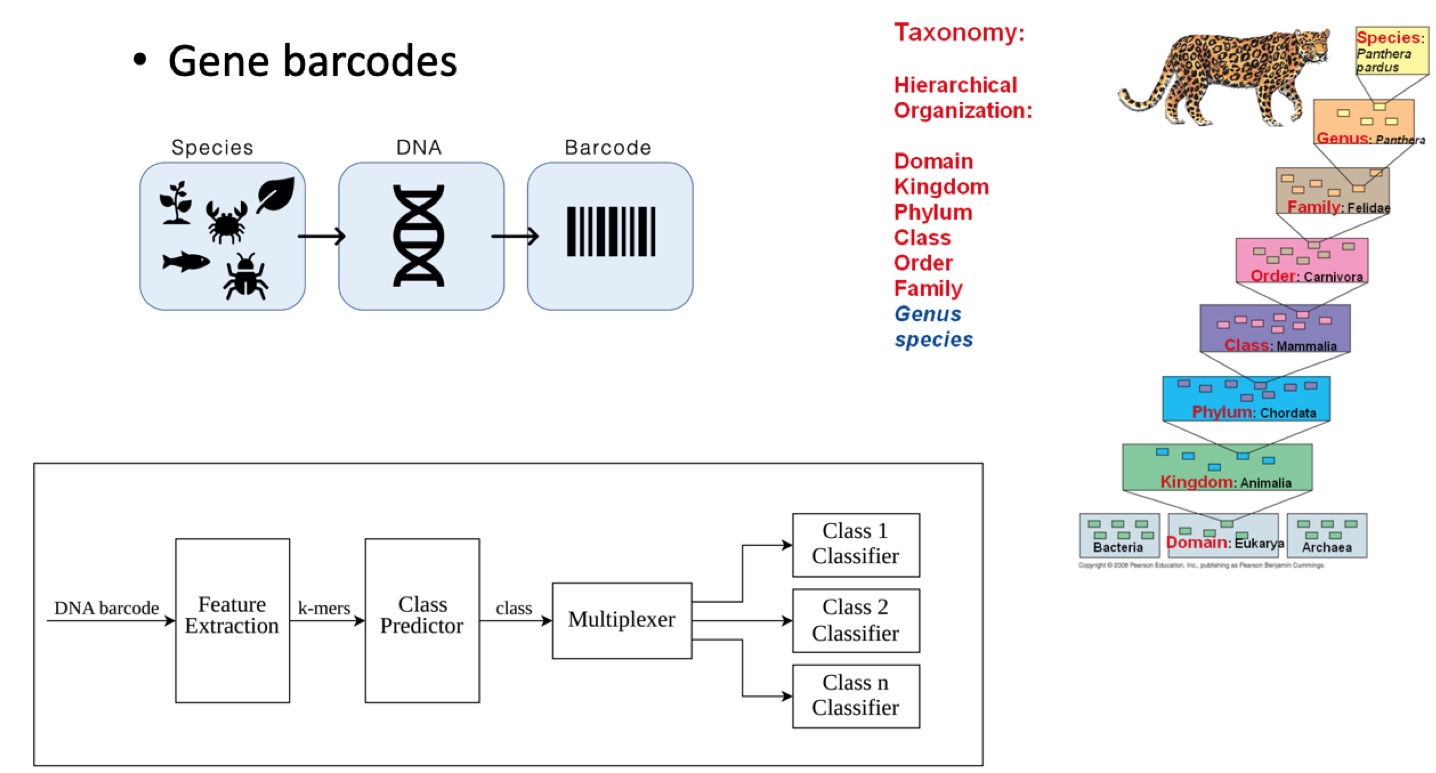
Scalable Taxonomy Classification
Various fields of applied biology (e.g., agriculture) depend on the classification of living creatures. However, many popular and highly accurate machine learning methods (e.g., Support Vector Machines) are not scalable to taxonomy settings where there is a large number of labels/classes. In this research, we developed a multi-level hierarchical classifier framework to automatically assign taxonomy labels to DNA sequences (Sohsah et al., 2020). We utilize an alignment-free approach called spectrum kernel method for feature extraction. We demonstrate that the proposed framework provides higher accuracy (i.e., 95%) than regular classifiers, and is scalable to taxonomy classification settings. Furthermore, we show that the proposed framework is more robust to mutations and noise in sequence data than the non-hierarchical classifiers.