Identification of Genomic Alterations Associated with Tumor Aggression in Tumor Subtypes
Abstract
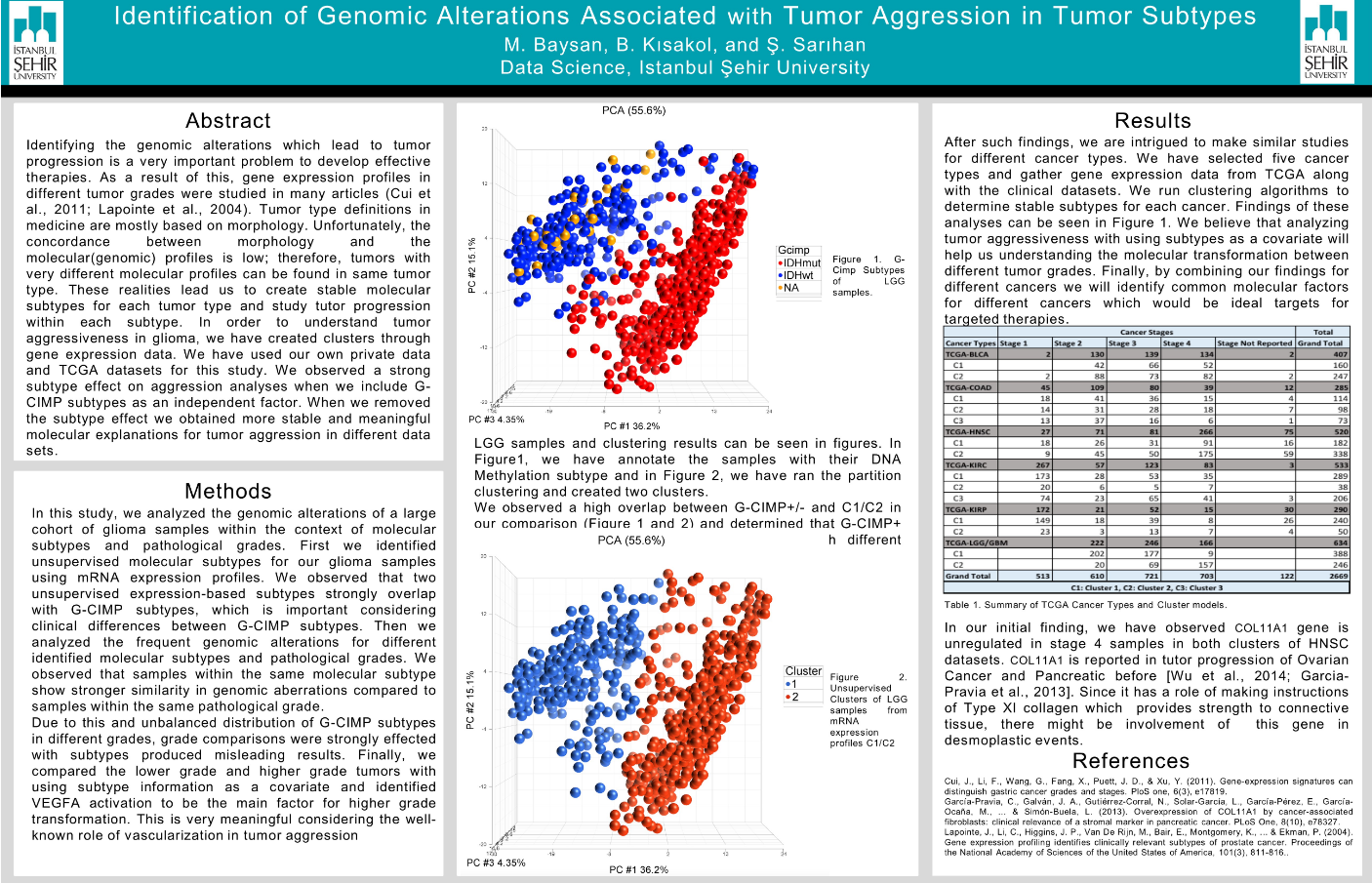
Identifying the genomic alterations which lead to tumor progression is a very important problem to develop effective therapies. As a result of this, gene expression profiles in different tumor grades were studied in many articles (Cui et al., 2011; Lapointe et al., 2004). Tumor type definitions in medicine are mostly based on morphology. Unfortunately, the concordance between morphology and the molecular(genomic) profiles is low; therefore, tumors with very different molecular profiles can be found in same tumor type. These realities lead us to create stable molecular subtypes for each tumor type and study tutor progression within each subtype. In order to understand tumor aggressiveness in glioma, we have created clusters through gene expression data. We have used our own private data and TCGA datasets for this study. We observed a strong subtype effect on aggression analyses when we include G-CIMP subtypes as an independent factor. When we removed the subtype effect we obtained more stable and meaningful molecular explanations for tumor aggression in different data sets. Methods In this study, we analyzed the genomic alterations of a large cohort of glioma samples within the context of molecular subtypes and pathological grades. First we identified unsupervised molecular subtypes for our glioma samples using mRNA expression profiles. We observed that two unsupervised expression-based subtypes strongly overlap with G-GIMP subtypes, which is important considering clinical differences between G-GIMP subtypes. Then we analyzed the frequent genomic alterations for different identified molecular subtypes and pathological grades. We observed that samples within the same molecular subtype show stronger similarity in genomic aberrations compared to samples within the same pathological grade. Due to this and unbalanced distribution of G-CIMP subtypes in different grades, grade comparisons were strongly effected with subtypes produced misleading results. Finally, we compared the lower grade and higher grade tumors with using subtype information as a covariate and identified VEGFA activation to be the main factor for higher grade transformation. This is very meaningful considering the well-known role of vascularization in tumor aggression.